Next-Generation Sequencing Based Yeast Two-Hybrid Screens
Summary
Protein-protein interactions are crucial for forming protein complexes to carry out designated protein functions and effectively underlie all biological functions and disease pathways. With the so-called "interactome" being the core to any living cell, mapping of protein-protein interactions (PPIs) is a major emphasis in biology and medicine. In addition, the molecular environment of a drug target and its potential interaction partners can be predictive of functional roles and unwanted side effects. We offer a novel Next-Generation Sequencing (NGS) Based Yeast Two-Hybrid (Y2H) screening service that overcomes false positives associated with traditional Y2H approaches and significant savings in time and cost.
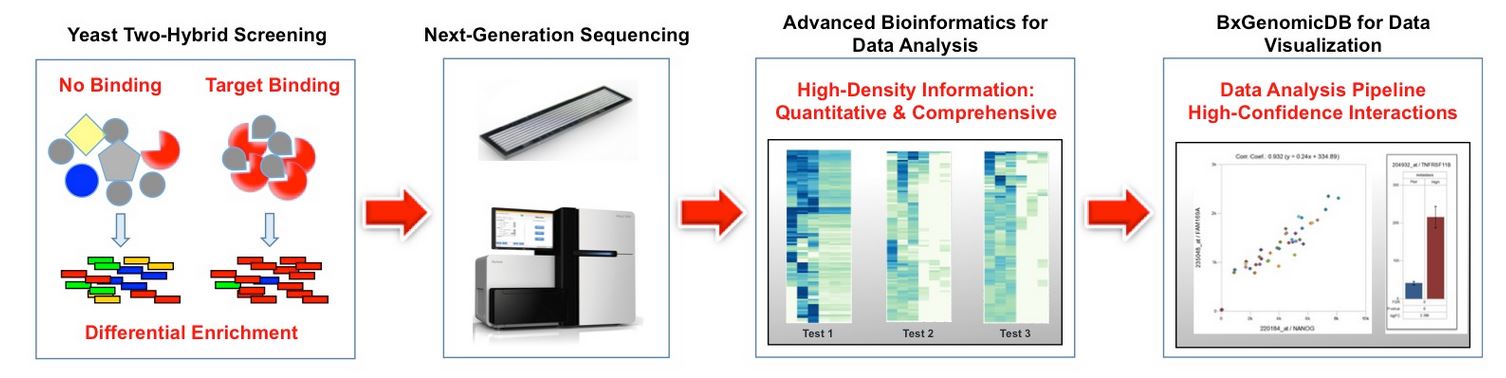
Key Features
- Higher throughput for better data coverage: NGS-Y2H yields at least 10-fold more data coverage of PPIs than the conventional approach.
- High quality data with low false positives: False positive generated by traditional Y2H can lead to unreliable datasets. Applying the NGS-Y2H procedure, confounding false-positives can be excluded a priori.
- Discover more novel interactions with advanced bioinformatics: Specialized bioinformatics algorithm for quick comparison across datasets and easy access to related experimental results.
Significant advantages: Higher quality screening data achieved upfront significantly simplifies downstream experimental design, shortens workflow, dramatically saves time and significantly lowers costs.
Applications:
- A complete direct protein-protein interaction (binary interaction) map of a target protein of interest
- Discover novel protein interaction partners under defined conditions
- Amino acid variants/mutations affecting protein-protein interaction
How to Order:
- Customer supply: protein target to be studied
- We deliver: Experimental design and execution, a comprehensive report and an optional web visualization of your data with advance bioinformatics tools
- For further inquiries and to discuss our services, please contact: BernhardS Suter, PhD. Y2H Screening Unit 1-510-558-0368(tel) 1-877-835-2534(fax) bernhard.suter@quintarabio.com
Next Generation Protein Interaction Screening (NGIS):
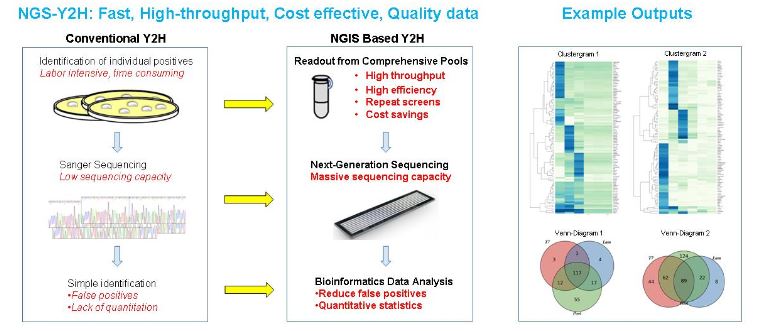
Protein-protein interactions (PPIs) underline all biological processes and disease phenotypes, and hence are paramount for biomedical and biological research. Yeast two-hybrid (Y2H) is the most widely used technology for the exploration of PPIs and is established for high-throughput screening. In most Y2H experiments, complex prey cDNA libraries are screened with a known bait protein. Individual yeast clones that contain interacting bait-prey combinations are then enriched using auxotrophic markers over millions of non-interacting combinations.
In conventional Y2H, selected preys are identified by Sanger sequencing. One-by-one identification of single cDNAs limits the efficiency of the Y2H methodology, and is a serious cost constraint for the identification of large numbers of PPIs. Moreover, the clonal identification of Y2H interactions is by no means a quantitative measurement and does not allow for systematic comparisons between screens. Lack of quantitation and slow identification of PPIs via Y2H is a sharp contrast to the gathering of gene expression and protein-DNA interaction data by RNA-Seq and Chip-Seq.
We developed a screening and scoring scheme for the identification of Y2H PPIs from comprehensive yeast pools using next-generation sequencing (NGS). Replacing conventional screening and Sanger sequencing of individual clones with a global selection and identification method has three major implications:
- First, a massive increase in sequencing capacity. This allows identification of all positives from Y2H screens at sharply reduced costs.
- Second, the increased sequencing capacity allows screening at low stringency to address every potential interaction from the cDNA library, i.e. maximum sensitivity.
- Third, NGS-Y2H allows the use of quantitative statistics to determine if a result is indeed bait-specific. Systematic controls allows a priori exclusion of false positive interactions, which used to be a major pitfall in conventional Y2H screens.


